Research objectives
Hollfelder Group research objectives

The Hollfelder group is interested in gaining a fundamental understanding of the principles responsible for molecular recognition processes in chemistry and biology.
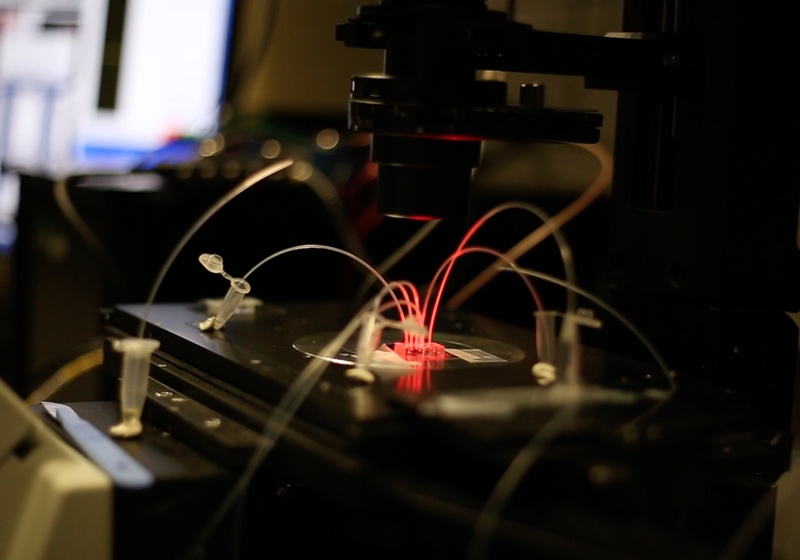
We use extreme miniaturisation of experiments (in picoliter droplets made in microfluidic devices) to answer chemical and biological questions
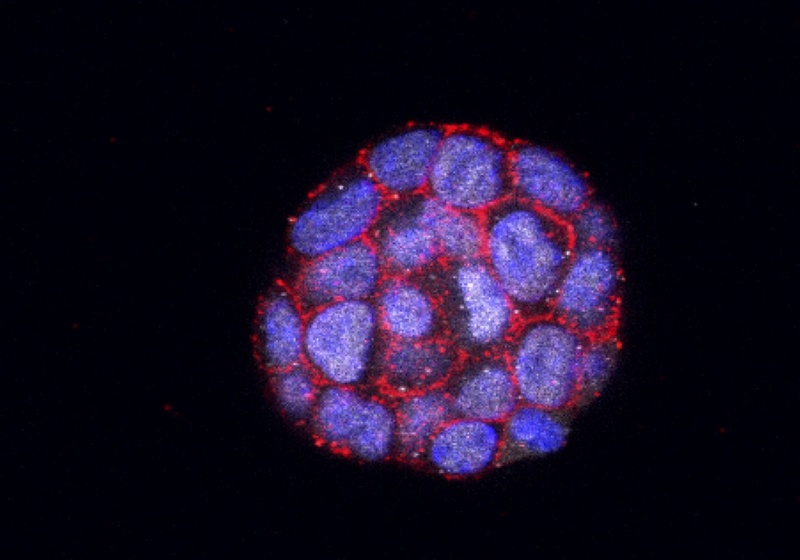
We encapsulate cells into microfluidic droplets to investigate single-cell transcriptomics, and understanding embryo development
Apply for a PhD
We have two open PhD positions in the EU Marie Curie Networks MetaExplore and On-TRACT (from summer 2026) - Deadline 12 January 2026

Hollfelder Group members
The Hollfelder group was started in 2001, we host Master's and PhD students, postdocs and visiting students.
